What do I do all day long?
I guess I already mentioned that I study the evolution of plants in tropical alpine ecosystems (: But what does that mean? What do I do? Besides going on field trips, visiting collaborators and conferences (see post about mobility), how does my work day look like? I usually spend the day in front of the computer.
To study the evolution of plants, we need some genetic information of the species we study. While in the past, it was limited to a few thousand base pairs that were variable enough to provide us with some information, but also not too variable that it was difficult ‘to get’ the region of the genome sequenced. These days, it became ‘easier’, we are able to sequence a few million base pairs. This means we have more information available to determine the relationships between the different species/samples. It also means that lab work got really complicated and expensive and that we need to be able to deal with all the data we receive. During my PhD, I was still working in the lab, generating the sequences I was interested in. Today, this is done by lab assistants who really know the knits and bits on how to do it best. One ‘drawback’ is, that we usually have to wait until we have 96 samples available that we would like to sequence - a particularity about how the sequencing works today.
So, these days, I choose the samples for sequencing, give them to our lab assistant and then just wait until I get a few hundred gigabytes of data from the sequencing facility. In order to deal with all the data, nowadays there are scripts available that are processing them and give us data back we can more easily work with. It usually takes a couple of days from the first scripts to the last ones to finish on a high performance cluster. In the beginning of the new sequencing technologies, we did not have these easy to use scripts and basically everyone had to come up with a homemade solution - often not very effective - but informatics is not part of a biology curriculum at most universities.
I was lucky and were able to attend a 10days workshop at the end of my PhD that introcuded me to programming in python, R and SQL. From there I started writing my own code and once you learnt the philosophy, it is also somewhat easy to transition from one programming language to another. It does not mean, my code is super efficient, but I can more easily accomplish tasks than without them (:
So, from all the sequences we get, I am calculating the species tree - the genetic relationships of my individual samples.
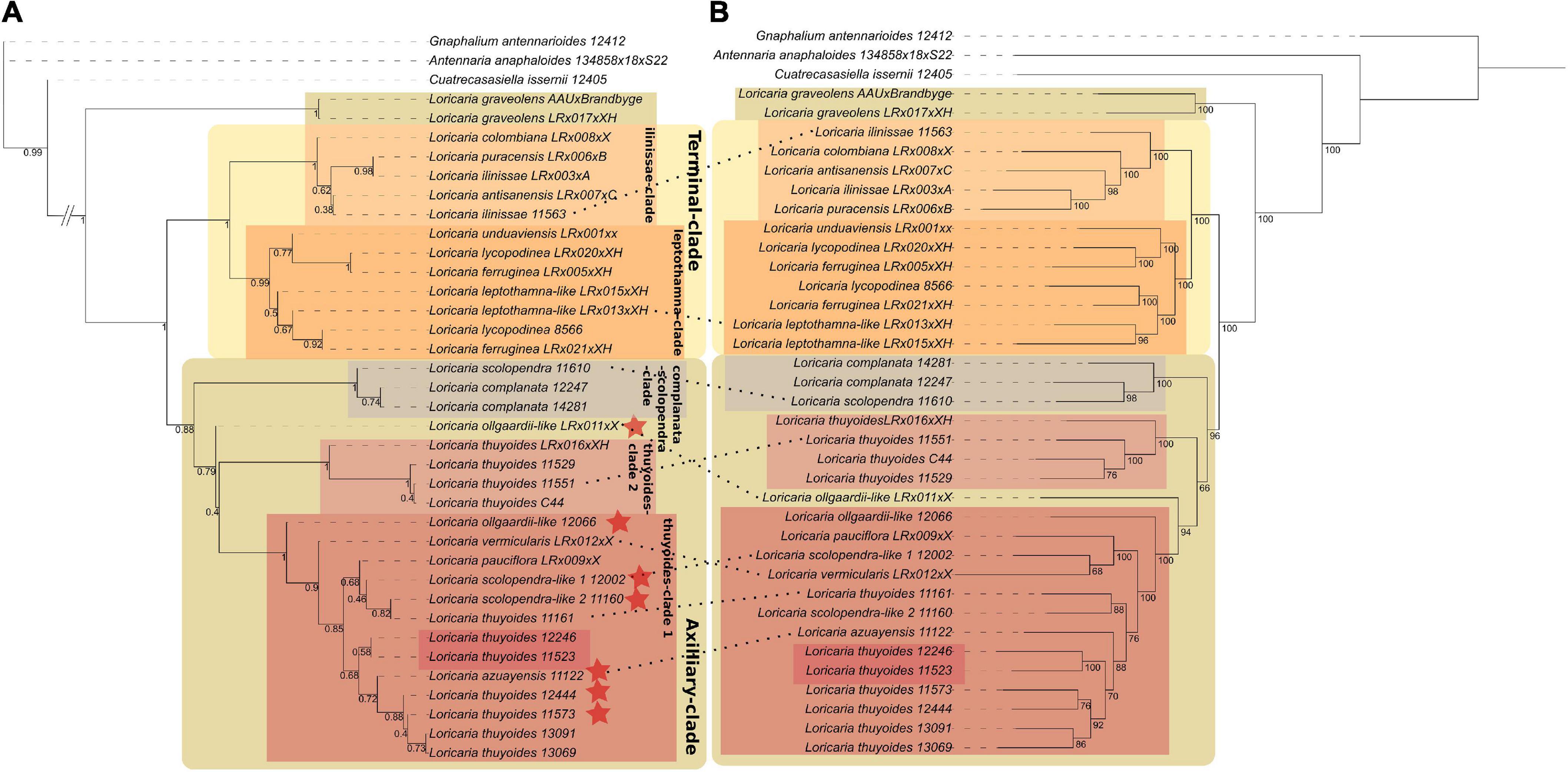 The species tree (A) of a study of mine published in ‘Frontiers in Plant Science’
The species tree (A) of a study of mine published in ‘Frontiers in Plant Science’
It is called species tree, as it is being calculated from all the underlying gene trees (sequences from the same genetic region are used to calculate those). They do not necessarily need to show the same relationship as the species tree, as certain gene regions might just evolve under a different scenario, due to processes such as hybridization. A phenomenom we also discovered in the above mentioned study - we found that species in the past must have hybridized. As such calculating the species tree is not as easy as it might sound, we often spend weeks to months doing so, as we need to filter out bad samples - due to low sequencing success, hybrids, etc.
Nevertheless, once we have our species tree, the real fun starts: we can date the whole tree using informations from fossils, which allows us to put the evolution of the species into a time frame.
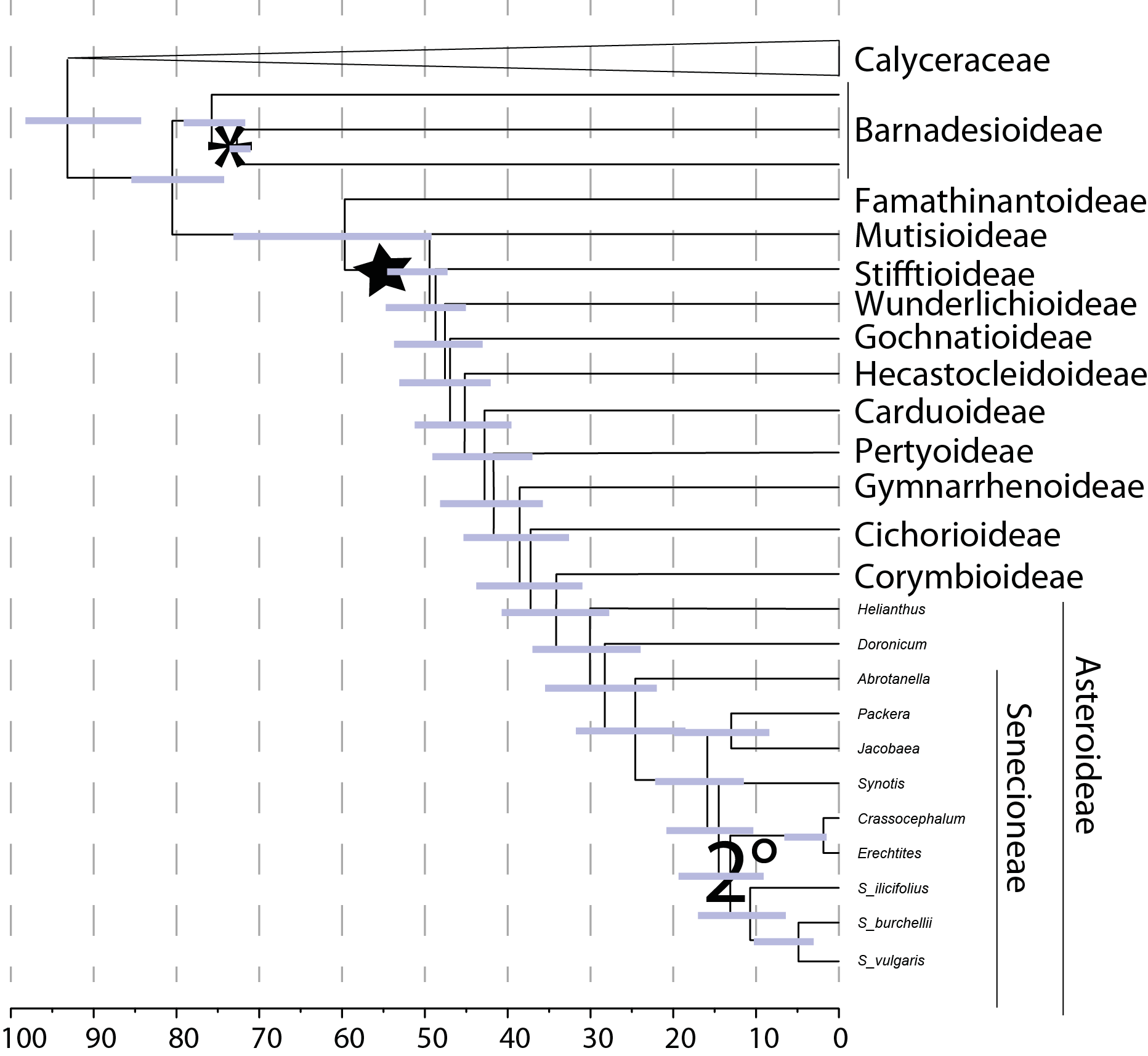 A dated phylogeny from my PhD project
A dated phylogeny from my PhD project
We can also determine where the most likely origin of all the species were, when there were bursts in diversification over time, if a certain morphological trait evolved only ones or in parallel, estimate if species were able to ‘meet’ geographically in the past and much more.
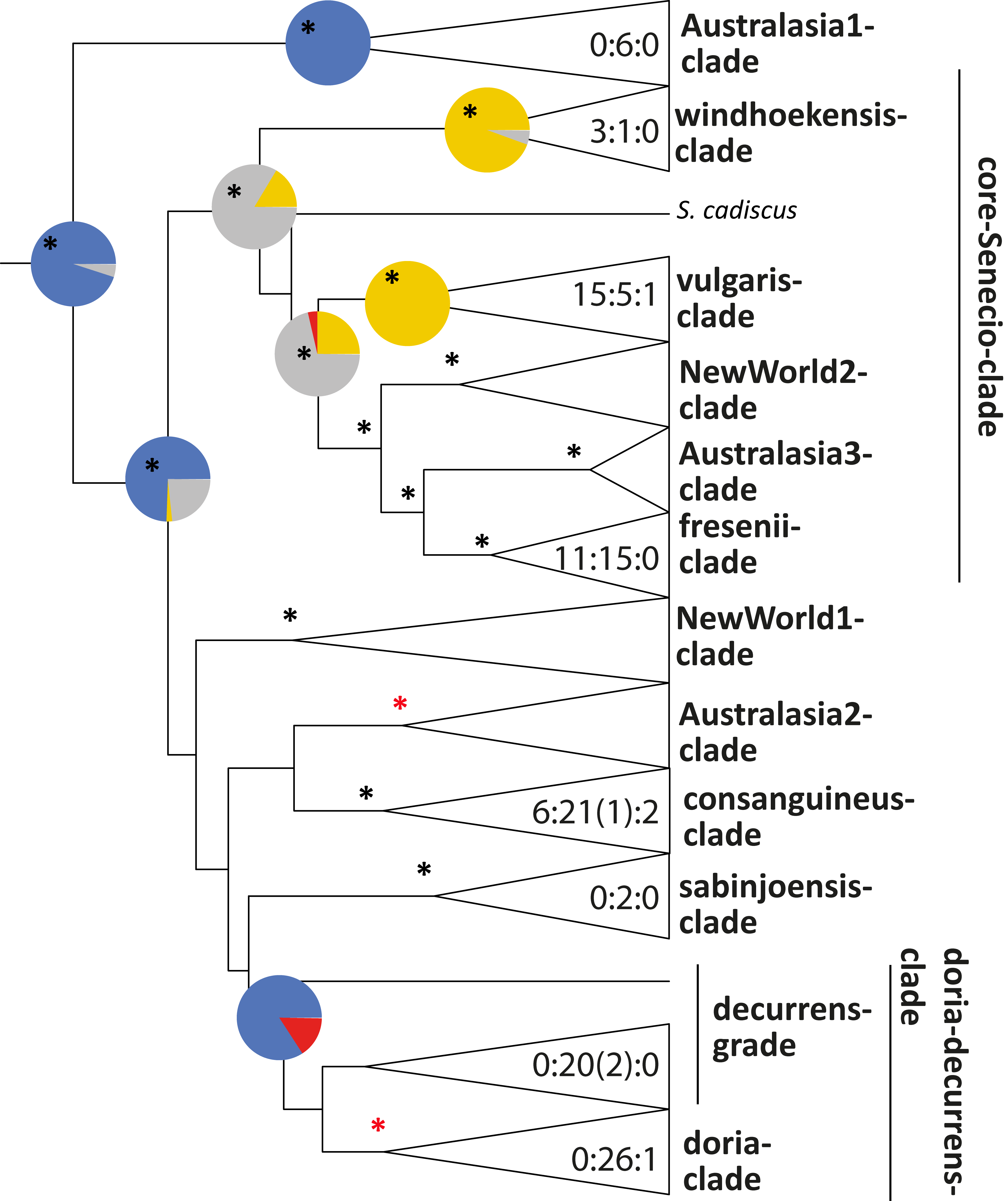 Another example from my PhD, trait reconstruction for the genus Senecio, perenniallity in blue, annual life history in yellow.
Another example from my PhD, trait reconstruction for the genus Senecio, perenniallity in blue, annual life history in yellow.
All these information are then used to gain a better understanding of the generation of biodiveristy. When and why do species diversify and form new species? Is every climate change event a trigger for extinction? Are the processes the same in different ecosystems and among different organisms? Why are some ecosystems more diverse than others? All these are questions we are trying to gain a better understanding of. As life is complex, it takes us some time to find answers and often those are: it depends (: but over time we are understanding more and more. For example, when I went to university it was still believed that hybridization does not lead to diversification, today we know it is a super important process for the diversification in flowering plants!
While at the moment, I spend most of my time in front of the computer doing analyses, my work is not just limited to that. I have to summarize the results into manucripts describing what I did, found out and how it fits into what we already know. On the other hand, journals are often contacting researchers to ‘review’ a manuscript of another author to ensure that it fits the scientific standards and the scope of the journal. We have internal seminars where we present our research and others can comment on it - a great opportunity to learn about others people research and broaden your own horizon. I have to write research grants trying to find money to fund my next project and/or apply for new positions (usually in my stage at academia, research contracts are for a maximum of two years - means we start looking for new opportunities as soon as we have started a new contract). Ah, and in my case, I am also teaching: I teach one class ‘Python for Biologists’, introducing the students to the basics of programming using biological examples. Supervision of master and PhD students is another thing we do. And of course, once in a while, we have the opportunity to actually leave the office and go on a field trip.