Development of workflows and tools for macroevolutionary research
I develop tools and workflows to increase reproducibility in research. Phylogenetic relationships of species are important if we want to study the diversification of lineages and the relations to ecological niches, biogeography and traits. I developed a tool to automatically update alignments based on custom sampling strategies. Colleagues and me also developed a python tool to easily and automatic update phylogenies using OpenTreeOfLife. Currently under development are workflows to analyze the climatic conditions of areas.
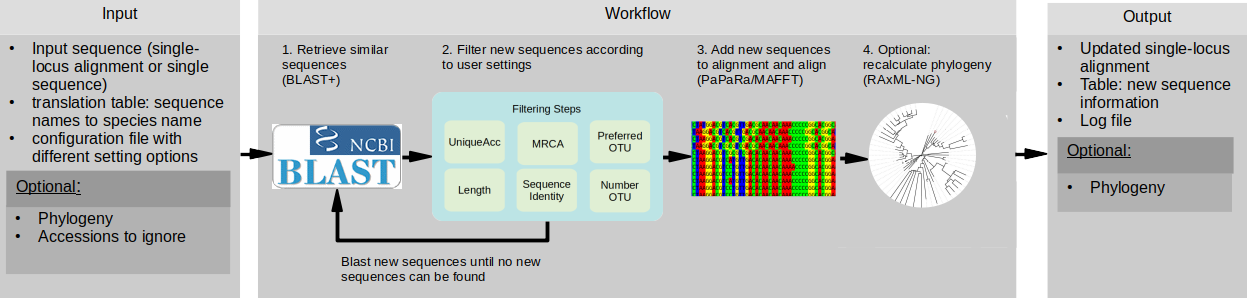 Workflow of PhylUp
Workflow of PhylUp
A new form of analysis method to understand the dynamics of diversification within the tropical alpine was developed during my Afroalpine dating project.
Besides, I have extensively using, testing and expanding usablity of a set of scripts developed by T. Fer and wrote a guideline on how to handle target enrichment data for recent radiations.
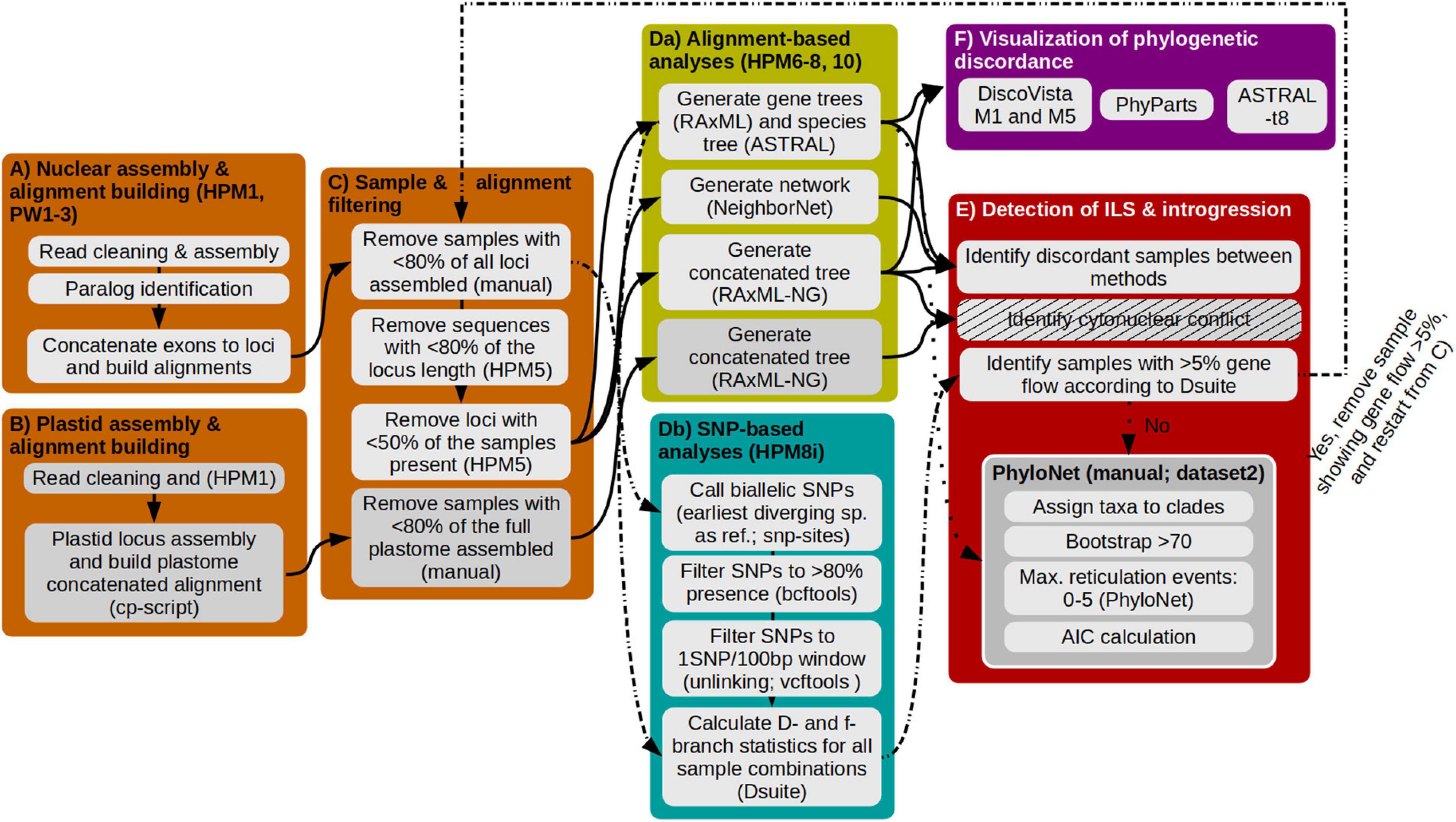 Workflow on how to handle genomic data of recent radiations
Workflow on how to handle genomic data of recent radiations